

The inverted repeat palindrome is also a sequence that reads the same forward and backward, but the forward and backward sequences are found in complementary DNA strands (i.e.Hint: In a double-stranded DNA or RNA molecule, a palindromic sequence is a type of nucleic acid sequence in which reading in a specific direction on one strand matches the reading in the opposite direction of the complementary strand.Ī restriction enzyme, also known as a restriction endonuclease or restrictase, is an enzyme that polymerizes DNA into pieces at or near restriction sites which are located inside the molecules.
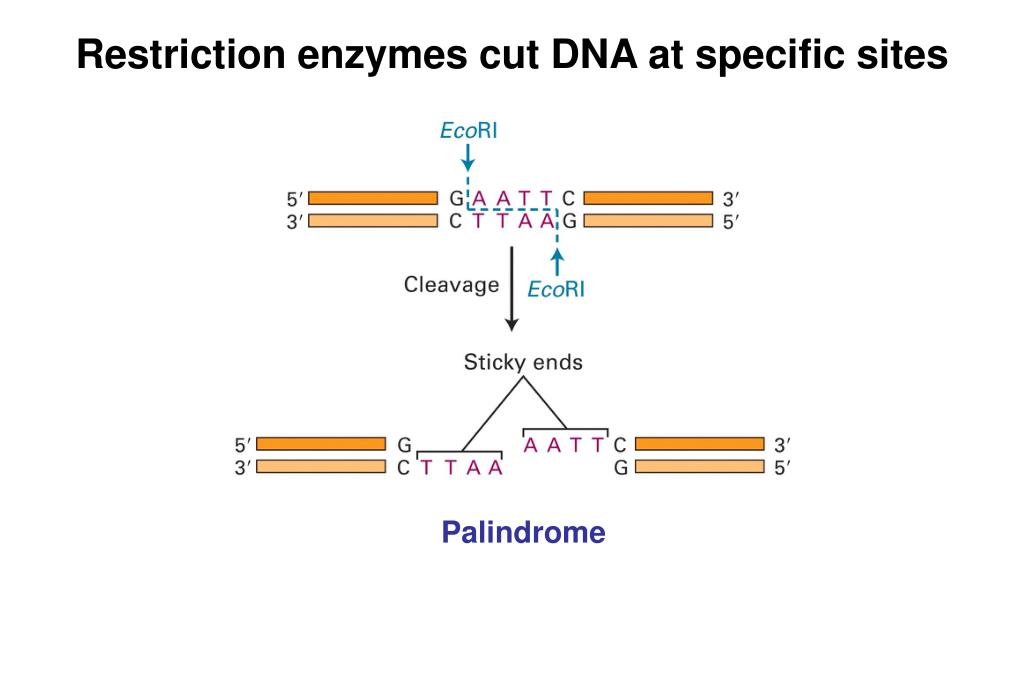
The mirror-like palindrome is similar to those found in ordinary text, in which a sequence reads the same forward and backward on a single strand of DNA, as in GTAATG. The recognition sequences of Type II Restriction Enzymes are palindromic, with two possible types of palindromic sequences. Ca 2 +, on the other hand, often acts as an inhibitor of Type II Restriction Enzymes. Almost all Type II Restriction Enzymes require divalent cations, usually Mg 2 +, as essential components of their catalytic sites. This cleavage generates reproducible DNA fragments, and predictable gel electrophoresis patterns, properties that have made these enzymes invaluable reagents for laboratory DNA manipulation and investigation. Type II Restriction Enzymes are a conglomeration of many different proteins that, by definition, have the common ability to cleave duplex DNA at a fixed position within, or close to, their recognition sequence. Neoschizomers, on the other hand, are often evolutionarily unrelated enzymes (e.g.EcoRII and MvaI). Isoschizomers that cut at the same position are frequently, but not always, evolutionarily drifted versions of the same enzyme (e.g. Isoschizomers that cut the same sequence at different positions are further termed ‘neoschizomers’ (neo = new). Restriction Enzymes that recognize the same DNA sequence, regardless of where they cut, are termed ‘isoschizomers’ (iso = equal skhizo = split). When there is more than one MTase, they are prefixed ‘M1.’, ‘M2.’, etc, if they are separate proteins or ‘M1∼M2.’ when they are joined.

The DNA-methyltransferases (MTases) that accompany restriction enzymes are named in the same way, and given the prefix ‘M.’. For example, the enzyme ‘HindIII’ was discovered in Haemophilus influenzae, serotype d, and is distinct from the HindI and HindII endonucleases also present within this bacterium. Roman numerals are used to specify different enzymes from the same organism. This is followed by letters and/or numbers identifying the isolate. The first letter of the enzyme refers to the genus of the organism and the second and third to the species. Restriction enzymes are named according to the taxonomy of the organism in which they were discovered.

Image from: Pingoud, A., Wilson, G.G., and Wende, W. Original Repository: Alan Mason Chesney Medical Archives, Daniel Nathans Collection. \): Hamilton Smith and Daniel Nathans at the Nobel Prize press conference, 12 October 1978 (reproduced with permission from Susie Fitzhugh).


 0 kommentar(er)
0 kommentar(er)
